Projects
Gene Discovery
Gene Discovery for Cerebral Palsy
- Status
- ongoing
- Rationale
- Cerebral palsy (CP) is the most common childhood-onset motor disability. A diverse set of risk factors contributes to CP. In approximately 20% of cases, there are no clear perinatal risk factors, suggesting the possibility of an underlying genetic disorder, including certain treatable inborn errors of metabolism. Therefore, understanding the genetic landscape of CP has implications for diagnosis as well as treatment.
- Description
- In this project, we are using whole exome sequencing to discover single gene disorders presenting as cerebral palsy, in instances where there are risk factors for CP, as well as those where there are no risk factors for CP.
- Acknowledgments
This study is part of the Children’s Rare Disease Cohorts (CRDC) Initiative, supporting gene discovery for different disease cohorts.
Neurodevelopmental Phenotyping
Neurobehavioral Phenotype of PTEN Hamartoma Tumor Syndrome
- Rationale
- PTEN Hamartoma Tumor Syndrome (PHTS) is a genetic disorder associated with a wide spectrum of systemic features and neurodevelopmental challenges, including intellectual disability (ID) and autism spectrum disorder (ASD). The neurodevelopmental challenges seen with PHTS can represent a significant source of impairment for affected individuals and families. Understanding the nature of these challenges can inform assessment and treatment of these symptoms.
- Description
-
We have characterized the neurodevelopmental phenotype of PHTS, comparing the neurodevelopmental profile of participants with PHTS and ASD to two other groups: participants with non-syndromic ASD with macrocephaly and participants with PHTS without ASD. We showed that PHTS is associated with a distinct neuropsychological profile suggestive of defects in frontal lobe systems, and that individuals with PHTS and ASD have more severe impairments in cognition, adaptive behavior, and sensory systems compared to individuals with PHTS without ASD. These findings help inform assessment and treatment of neurobehavioral symptoms associated with PHTS, with and without ASD.
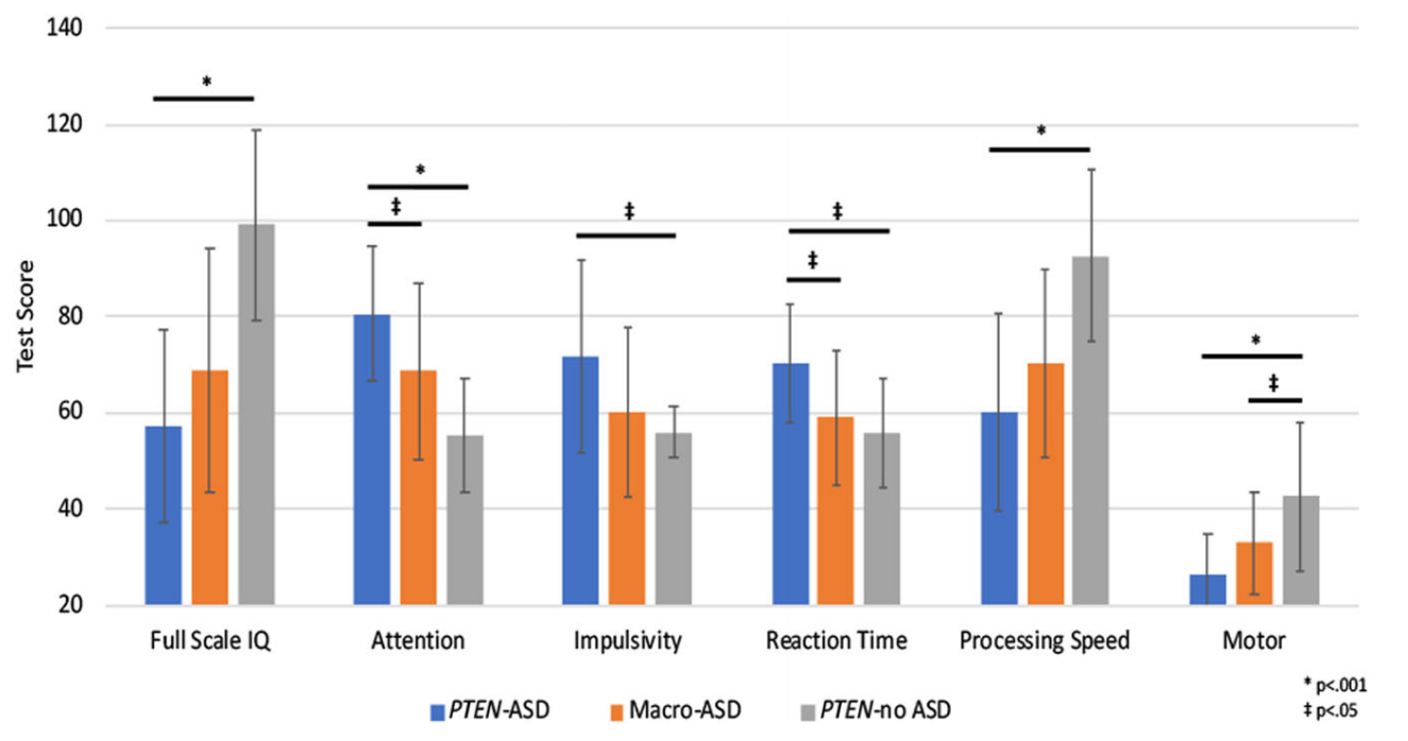 Graph showing impairments in various neurocognitive domains experienced by individuals with PHTS, with and without ASD [Pubmed]
Graph showing impairments in various neurocognitive domains experienced by individuals with PHTS, with and without ASD [Pubmed] - Acknowledgments
Collaborators at Cleveland Clinic (including Dr. Robyn Busch and Dr. Charis Eng) spearheaded this work. This study used data from the NIH-sponsored Developmental Synaptopathies Consortium, led by Dr. Mustafa Sahin.
- Publications
-
-
Neurobehavioral Phenotype of Autism Spectrum Disorder Associated with Germline Heterozygous Mutations in PTEN
Transl Psychiatry. 2019 Oct 8; 9 (1): 253
[Pubmed]
-
Neurobehavioral Phenotype of Autism Spectrum Disorder Associated with Germline Heterozygous Mutations in PTEN
Biomarker Identification
Neuroimaging Biomarkers of Cognition and Behavior in Phelan-McDermid Syndrome
- Rationale
- Autism spectrum disorder (ASD) and intellectual disability (ID) may represent final common pathways in single gene disorders associated with a high prevalence of both of these conditions. Therefore, understanding the mechanisms of ASD and ID through the lens of specific genetic disorders may lead to insight into ASD and ID in general. Phelan McDermid Syndrome (PMS) is one such example of a monogenic model of ASD and ID.
- Description
-
We have investigated neuroimaging biomarkers in PMS. We conducted volumetric analysis on the brain MRIs of individuals with PMS. We found that compared to healthy controls, individuals with PMS have decreased volumes of the striatum and thalamus, structures which may play a role in repetitive behaviors. Using diffusion tensor imaging (DTI) analysis, we also showed that PMS is associated with decreased integrity of the uncinate fasciculus (UF). The UF is implicated in social and emotional interaction, and abnormalities in this tract may underlie some of the deficits seen in the PMS population. However, continued research needs to be done with larger sample sizes in order to replicate and build on these studies. These investigations are a starting point in helping understand how changes in the brain in PMS lead to ASD, ID, and other neurodevelopmental challenges.
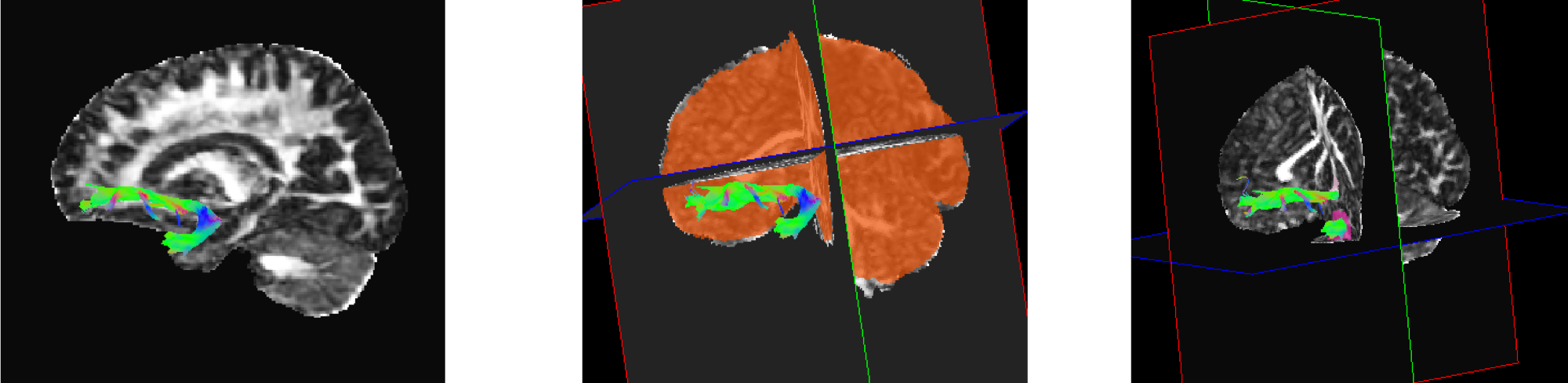 Example images of the UF in two different patients with PMS. [Pubmed]
Example images of the UF in two different patients with PMS. [Pubmed] - Acknowledgments
These studies used data from the NIH-sponsored Developmental Synaptopathies Consortium, led by Dr. Mustafa Sahin, as well as computational resources and expertise from the Computational Radiology Laboratory, led by Dr. Simon Warfield.
- Publications
-
-
Volumetric Analysis of the Basal Ganglia and Cerebellar Structures in Patients with Phelan-McDermid Syndrome
Pediatr Neurol. 2019 Jan 1: 37-43
[Pubmed] -
Diffusion Tensor Imaging Abnormalities in the Uncinate Fasciculus and Inferior Longitudinal Fasciculus in Phelan-McDermid Syndrome
Pediatr Neurol. 2020 May: 24-31
[Pubmed]
-
Volumetric Analysis of the Basal Ganglia and Cerebellar Structures in Patients with Phelan-McDermid Syndrome
Clinical Practice Changes
Diagnostic Yield and Clinical Utility of Exome Sequencing for Neurodevelopmental Disorders
- Rationale
- Individuals with neurodevelopmental disorders (NDDs) often undergo a diagnostic odyssey searching for a genetic cause. This can not only lead to significant distress but also reduce opportunities for potential intervention during critical periods in brain development. Understanding the diagnostic yield of different genetic tests for NDDs can help guide clinicians on test selection, thereby reducing the time in this diagnostic odyssey.
- Description
-
We conducted a retrospective study that evaluated the diagnostic yield of exome sequencing when applied to the neurodevelopmental disability population and detailed how the results of exome sequencing changed management for individuals with a genetic diagnosis. With a team of international collaborators, we also performed a meta-analysis as the basis for a consensus statement suggesting that exome sequencing serve as a first line test for NDDs like autism spectrum disorder (ASD) and intellectual disability (ID).
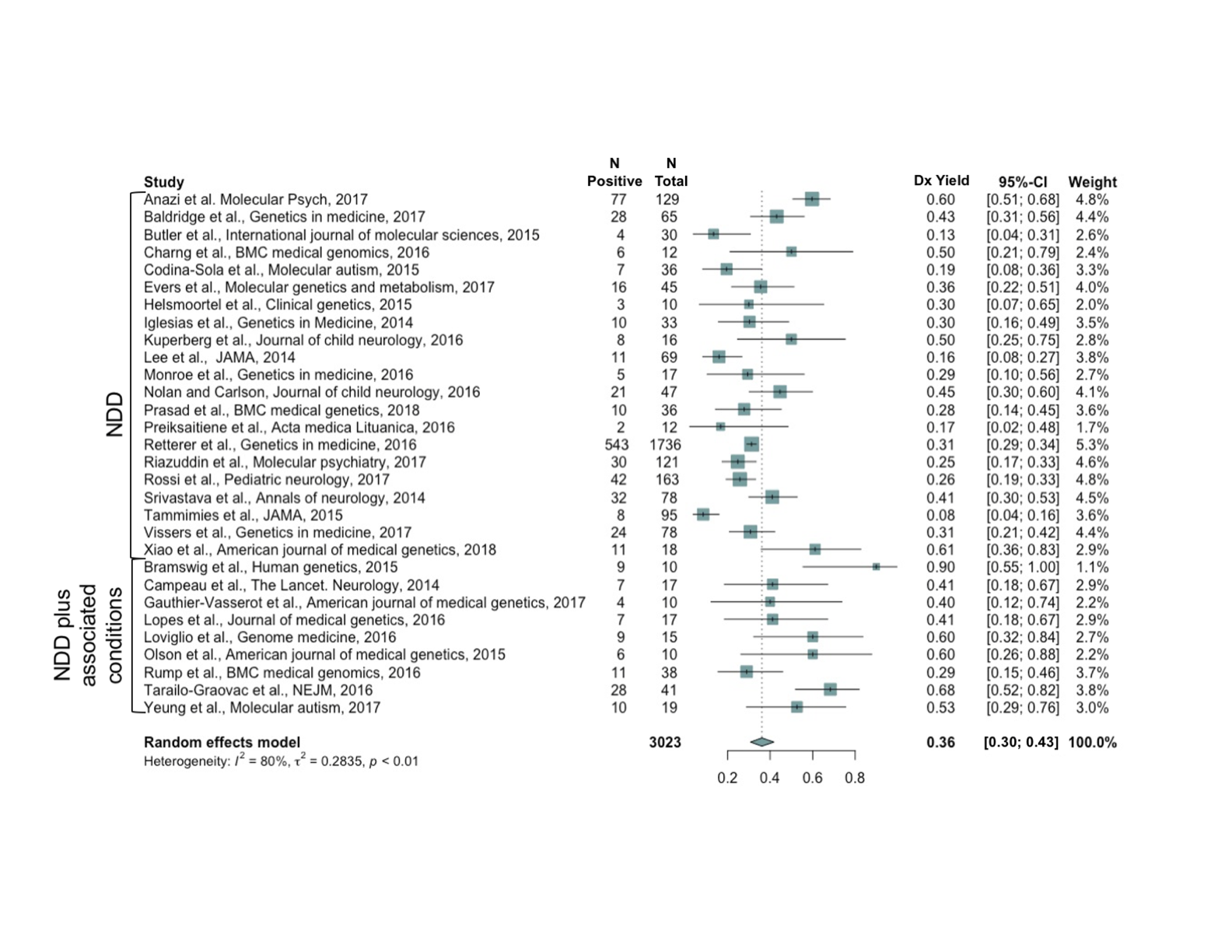 Tree plot showing meta-analysis of studies examining diagnostic yield of exome sequencing for neurodevelopmental disorders [Pubmed]
Tree plot showing meta-analysis of studies examining diagnostic yield of exome sequencing for neurodevelopmental disorders [Pubmed] - Acknowledgments
Dr. Mustafa Sahin and Dr. David Miller led the effort to develop the consensus statement, which involved contributions from multiple international collaborators.
- Publications
-
-
Clinical Whole Exome Sequencing in Child Neurology Practice
Ann Neurol. 2014 Oct; 76 (4): 473-83
[Pubmed] -
Meta-analysis and Multidisciplinary Consensus Statement / Exome Sequencing is a First-tier Clinical Diagnostic Test for Individuals with Neurodevelopmental Disorders
Genet Med. 2019 Nov; 21 (11): 2413-2421
[Pubmed]
-
Clinical Whole Exome Sequencing in Child Neurology Practice